

Advances in the life sciences have resulted in an increase in the volume of data

“My goal is to create an immense online life sciences dictionary containing information on measured values, genome sequences, gene expressions, metabolic pathways and knowledge of agronomic traits in crops.” says Associate Professor Yano.
The volume of data handled within the field of life sciences is increasing greatly, and extensive data analysis cannot be carried out even with large-scale computers. We now hear the term “big data” in the field of life sciences more often. To completely perform a large-scale computer analysis with such large genomic data, a new method is required, and I am among those engaged in establishing such methods.
Can computers provide knowledge and new findings from large text data?
Genome sequencing is just a base sequence decoding. Therefore, we are not checking if each one of several tens of thousands of genes on a genome sequence actually exists by experiments. At this point, the computer is simply predicting what kinds of genes are existing on which certain regions of the genomic sequence, and what kinds of activities they are performing. Although genome sequencing was an important milestone for the development of life sciences, how we can obtain useful information using this data depends on the future research. The current situation is as if we had just obtained a blank map.After the determination of genome sequences, we predict genes on the genome and their biological functions by using current sequence databases. However, in general, the reliability and validity of data provided from databases varies. For example, even if it has been reported that a certain gene works to form the eyes of a drosophila fly, we cannot eliminate the possibility that this information is incorrect. We currently have no system of verifying an assertion. With some publications, we must verify functions of the genes.
To provide information on biological functions of genes, the curators read papers and store the information into a database. Although this manual curation is very accurate, it entails time and high labor costs. In my laboratory, we are engaged in utilization of natural language processing (NLP). With approaches with NLP, computers treat large-scale text information from publication data, and try to mine knowledge and new findings. If we can enable the computer read a paper, understand and deposit new additional information into the database server, a highly-reliable knowledge information database can be constructed automatically. Although studies concerning NLP have made considerable progress, the computer is not yet able to perform processing as well as humans do. The major challenge we are facing now is to make the computer understand the context of the papers, and deposit new knowledge into the database.

I also participate in research to improve breeding efficiencies by utilizing genetic information. This research is aimed at the production of a biofuel at a low price.
Understanding an organism as a system
Another keyword is systems biology. This is an even newer field of research with the purpose of understanding organisms as a system. Under initial genome sequencing era, most researchers considered genome sequence data could provide key insights into more complicated mechanisms in organisms. However, it is quite insufficient only with genome information. We must collect other omics data such as gene and protein expressions, differences in DNA sequences among individuals, and so on. We consider organisms, organs and cells act like a system. We are eager to understand how the system work under rigorous but flexible processes. In plain terms, the theme of my research is to discover the dynamic characteristics of the system through an integrated analysis of enzymes, genes, proteins, etc.What determines species-specific traits?
Molecular biological experiments, such as next generation sequencing (NGS) technology has been rapidly advancing. Even human personal genomes can be shortly determined by the current NGS platforms. In addition, NGS and microarray experiments can quickly provide large-scale gene expression data.On the other hand, due to an excessive increase in the amount of experimental data, we are overwhelmed, feeling as though we are sitting on a gold mine without a pickaxe. The large-scale data make computational analysis difficult due to short of computer memory and/or long calculation time. Actually, large-scale gene expression analysis sometimes cannot be performed due to the large amount of data. To complete large-scale computational analysis, it has been frequently done to reduce the data size for the further analysis. For example, by removing genes which don’t show significant differences in expression levels among samples, the data size decreases as expression analysis can be performed. However, this approach takes risk to eliminate genes for the further analysis, which actually concern with important trait such as crop yield, human disease, and so on. Our challenge is to develop a new method which allows us to quickly perform large-scale analysis with general computer system.
Even if the large-scale computer analysis completes, the data from the analysis are also too large-scaled to be quickly interpreted. Therefore, we have developed software with graphical viewer which helps biologists to understand the large-scale data from analysis. The viewer summarizes the results in graphical 3D space. Users can exploit the viewer to discover genes with specific expression patterns.
Since our software can be executed even on laptop computers, it suggests new great potential for a remarkable explosion of biological discovery by many of biologists, who could not have performed large-scale analysis by themselves to date. I consider the approaches which can be easily and quickly executed within low-cost computers such as laptop PCs in general laboratories, large-scale computers are not only for bioinformaticians, it facilitates to discover novel genes useful for human health, crop yield and so on.
Furthermore, by using a gene expression network that easily recognizes genes with similar expression patterns, genes of interest can be quickly identified. In addition, if heterogeneous networks can be compared and analyzed, genes capable of providing individuality to each species can be identified. What endow rice specific characters in rice? – I want to discover something that exists only in rice among the genomes.

Developing experts in bioinformatics is another challenge
I believe it would be ideal if researchers will be able to analyze their own data themselves even in the traditional agricultural studies. Entrusting data handling to someone else takes time to discuss the aims and approaches of the research. It is impossible for another person, including bioinformaticians, to completely understand your experimental designs, approaches and goals. So, bioinfomaticians who analyze your data may sometimes execute the analysis in a wrong way. In addition, it is difficult for biologist to recognize that the results are obtained by inappropriate ways. When you trust the data, you will publish the paper on the basis of the results. This status in which biologists don’t analyze their own data by themselves will not lead to the development of the life sciences. The current biologists must have biological knowledge and computer skills to handle big data to derive a conclusion in life sciences. Food production is an important national strategy. To provide a foundation for sustainable agriculture, bioinformatics approaches are essential to make the most of extensive genetic resources in the plant kingdom. However, bioinformaticians are very few in Japan since the educational system is still insufficient. We eagerly require an enhanced educational infrastructure for bioinformatics researches.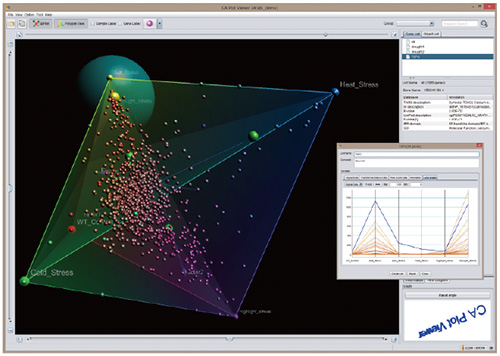
An image using 3D software developed by Associate Professor Yano. Comparison of the gene expression patterns is easy by the graphical viewer.
Profile
Associate Professor, Department of Life Sciences, School of AgricultureResearch interest:Applied Genomic Science, Bioinformatics, Systems Biology
Researcher, Kazusa DNA Research Institute; Project Research Associate, University of Tokyo
Associate Professor, School of Agriculture, Meiji University, 2011
Ph.D. in Agriculture, Kyoto University










